Artificial intelligence technology creates proteins from scratch
Brain area necessary for fluid intelligence identified / Will machine learning help us find extraterrestrial life?
The experiment demonstrates that natural language processing can learn at least some underlying biological principles, despite being designed to read and write text. ProGen is an artificial intelligence programme created by Salesforce Research that uses next-token prediction to assemble amino acid sequences into artificial proteins.
According to scientists, the new technology could become more potent than directed evolution, the protein design technology that won the Nobel Prize, and it will revitalise the 50-year-old field of protein engineering by accelerating the development of new proteins that can be used for almost anything, from therapeutics to degrading plastic.
"Artificial designs perform significantly better than designs inspired by the evolutionary process," said James Fraser, PhD, professor of bioengineering and therapeutic sciences at the UCSF School of Pharmacy and co-author of the study published in Nature Biotechnology on January 26.
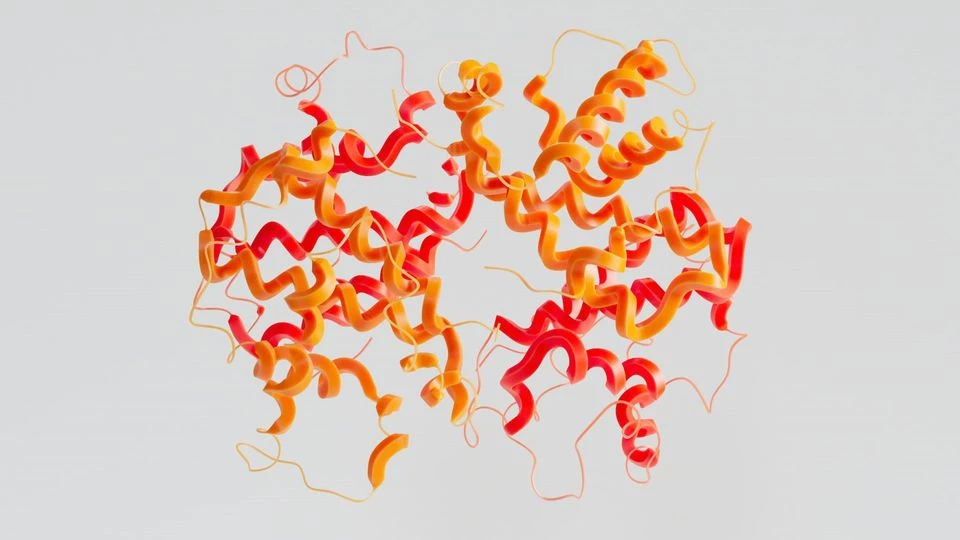
Artificial intelligence technology creates proteins from scratch.
"The language model incorporates aspects of evolution, but in a manner distinct from the typical evolutionary process," said Fraser. "We can now adjust the generation of these properties for particular effects. For instance, an enzyme that is extremely thermostable, prefers acidic conditions, or does not interact with other proteins."
To create the model, scientists simply fed the amino acid sequences of 280 million different types of proteins to a machine learning model and let it digest the data for two weeks. Then, the model was fine-tuned by priming it with 56,000 sequences from five lysozyme families and contextual information about these proteins.
The model quickly generated a million sequences, and the research team chose 100 to test based on how closely they resembled the sequences of natural proteins, as well as how naturalistic the underlying "grammar" and "semantics" of the AI proteins' amino acid sequences were.
Out of the first 100 proteins screened in vitro by Tierra Biosciences, the team created five artificial proteins to test in cells and compared their activity to hen egg white lysozyme, an enzyme found in the whites of chicken eggs (HEWL). Similar lysozymes are present in human tears, saliva, and milk, where they function as antibacterial and antifungal agents.
Two of the artificial enzymes were able to degrade bacterial cell walls with comparable activity to HEWL, but their sequences were only about 18% identical. Approximately 90% and 70% of the two sequences were identical to any known protein.
A single mutation in a natural protein can render it inactive, but in a separate round of screening, the team discovered that AI-generated enzymes were active even when only 31.4% of their sequence resembled a known natural protein.
Even from studying the raw sequence data, the AI was able to figure out how the enzymes should be formed. Using X-ray crystallography, the atomic structures of the artificial proteins appeared as expected, despite the sequences being unprecedented. In the year 2020, Salesforce Research created ProGen based on a type of natural language programming that their researchers had initially created to generate English language text.
Based on their previous research, they knew that the AI system could teach itself grammar, word meaning, and other underlying rules that make for well-composed writing.
Nikhil Naik, PhD, Director of AI Research at Salesforce Research and the paper's senior author, stated, "When you train sequence-based models with a large amount of data, they are very effective at learning structure and rules." They gain knowledge of co-occurrence and compositionality.
With proteins, the design options were nearly infinite. Lysozymes are small compared to other proteins, containing up to 300 amino acids. However, since there are 20 possible amino acids, there are 20300 possible combinations. That is greater than multiplying the number of grains of sand on Earth by the number of atoms in the universe by the number of humans who have ever lived. Given the infinite possibilities, it's remarkable that the model can generate functional enzymes so easily.
Ali Madani, PhD, founder of Profluent Bio, former research scientist at Salesforce Research, and the paper's first author, stated, "The ability to generate functional proteins from scratch out-of-the-box demonstrates that a new era of protein design is beginning," This versatile new tool is now available to protein engineers, and we eagerly await therapeutic applications.
Journal Reference: Ali Madani, Ben Krause, Eric R. Greene, Subu Subramanian, Benjamin P. Mohr, James M. Holton, Jose Luis Olmos, Caiming Xiong, Zachary Z. Sun, Richard Socher, James S. Fraser, Nikhil Naik. Large language models generate functional protein sequences across diverse families. Nature Biotechnology, 2023; DOI: 10.1038/s41587-022-01618-2
End of content
Không có tin nào tiếp theo
